Blue Genes: How Genome Research Can Boost Global Aquaculture
| Thu, 19 Mar 2020 - 16:08
Despite a number of challenges that still need to be overcome, genome sequencing has a growing range of practical applications in aquaculture and, as costs come down, it is going to be increasingly widely used.
It hardly seems like 20 years ago when the Human Genome Project announced that it had sequenced “a working draft” of the human genome. Genome sequencing has become faster and cheaper since the project began and we are now able to get our individual genomes sequenced by commercial labs. But what is genome sequencing and what, if anything, does it mean for aquaculture?
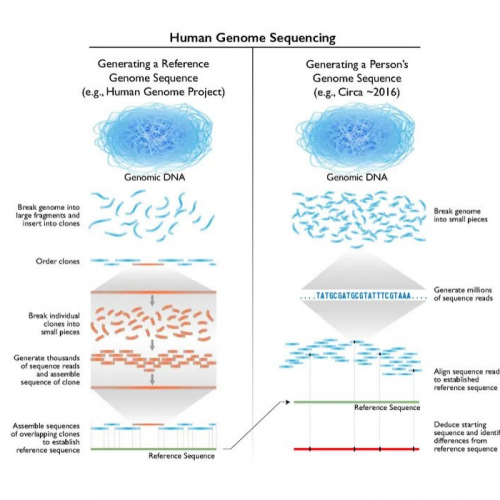
How genome sequencing was achieved for the Human Genome Project. Technologies vary, but the principle is the same for sequencing the genomes of aquatic species too
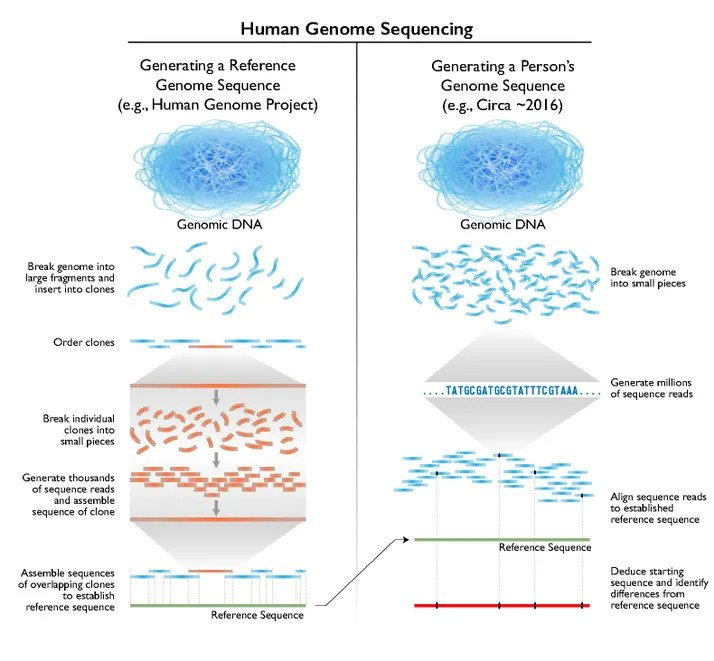
© Credit National Human Genome Research Institute
Background
The molecule DNA is often characterised as containing “the building blocks of life”, with its long chemical sequences forming the genes that control so many aspects of our physiology. Most of our genes are no different from one person to the next, but within this uniformity there are small variations which give rise to some of the characteristics that make us unique, like eye colour. The full suite of an individual’s genes is known as their genome. Put simply, genome sequencing is a way for us to read the genes in a genome.
Building up a library of genomes, we can find where there are variations between individuals, and unravel how these variations affect characteristics. We can also better understand how genes interact with each other, and even with the environment, to produce variations in characteristics – such as how temperature can influence the size of fish. We can also compare genomes from one species to the next, seeing if the same genes control the same characteristics, or if similar variations or mutations lead to similar outcomes.
Although currently less prevalent in aquaculture than in terrestrial farming, embracing genetics is not new. Traditional selective breeding methods rely on us looking at an individual’s phenotype – how its genes are expressed in its environment. If we see an individual grows particularly large, for example, we may want to use that individual for breeding larger offspring. In fact, genetics underlies all sorts of characteristics of interest to aquaculturists.
Just over a year ago, Dr Hugues de Verdal, based at CIRAD in France, published his work demonstrating that feed efficiency is a heritable trait in Genetically Improved Farmed Tilapia (GIFT). In fact, de Verdal estimates that genetics explains around 30 percent of a tilapia’s feeding efficiency, which is no small amount. Because the trait is heritable – driven by the genes of the individuals – it is possible to selectively breed the tilapia to promote feeding efficiency.

Dr Hugues de Verdal's research demonstrated that feed efficiency is a heritable trait in Genetically Improved Farmed Tilapia (GIFT)
© Hugues de Verdal
De Verdal’s work is among the first to identify the genetic underpinnings of feed efficiency in aquatic species. Although he did not use sequencing in this study, de Verdal believes that embracing such techniques has substantial benefits, namely in terms of precision and predictive ability.
“You will have more information about the genes given by the parents, so it is more accurate to use genomic techniques to estimate the heritability [of a trait] and to estimate the breeding values of each fish,” he explains.
Sometimes the genetic basis of characteristics is relatively easy to isolate. Take antifreeze protein genes, for example, which allow some species to survive in sub-zero temperatures. Experimental antifreeze-protein-gene transfer from winter flounder into fertilised Atlantic salmon eggs was part of the early stages of transgenic salmon research.
Most traits that aquaculturists desire, however, do not come from a single gene. De Verdal suspects that feed efficiency arises from a suite of genes.
“We are quite sure there is no one gene which is leading all aspects of feed efficiency because it could come from a lot of different things, such as assimilation of nutrients, potentially the size of the gastrointestinal tract and expiration,” he explains. “It's probably really complex.”
Genome sequencing helps untangle such complexities. The potential applications for aquaculture are many. For the hatchery operator aiming for a monosex population, understanding how genes and the environment interact on a genotypic rather than phenotypic level can, for example, precisely guide choice of temperature, which influences sex in some species. For breeders, advanced selective breeding techniques allow for efficient and precise development of strains specifically cultivated for a range of farming conditions. It may even be possible to manipulate genes that control characteristics like disease resiliency. Some researchers have even started sequencing pathogen genomes which, among other things, can help identify transmission pathways, helping improve disease management. For those who use wild broodstock, understanding the genome of the wild population can not only help manage the broodstock fishery, but also ensure that you haven’t accidentally picked up a species that looks like the one you want, but actually is something else.
This is just a small sample of the potential applications, the number of which will invariably rise as aquatic-species genomic research becomes more commonplace.

A machine used to sequence genomes
© Kenneth Rodrigues
Challenges
As with any technological application, there are challenges, some of which are unique to aquatic species. As Professor Kyall Zenger at James Cook University in Australia highlights, this includes the relatively high levels of polysaccharides found in animals such as crustaceans and shrimps.
“Polysaccharides are a complex compound used for defence mechanisms and other biological processes, but they inhibit sequencing,” Zenger explains. “It’s not as simple as extracting [genomes] from a terrestrial animal that has been routinely sequenced.” In fact, for most aquaculture species, there is no baseline genome sequence from which researchers can begin working. Both Zenger and de Verdal point out that building a baseline of any species can mean sequencing the genome of thousands of individuals.
A further challenge relates to the highly repetitive genomes of aquatic species – and their high rates of mutations. When a genome is sequenced, we do not get a single, neatly ordered output. Genomes are sequenced in small parts, which are then put back together in the correct order. “The more complex the genome and the more repetitive it is, the more difficult it is to put back together because the [sequencing] software gets confused, so to speak,” Zenger explains.
Zenger notes that genomic technology applications on aquatic species are still in the early stages of development and is confident that the technological challenges – and the lack of baseline sequences – will be overcome.
For aquaculturists eager to start using genome sequencing directly in their farms or hatcheries, there is some bad news.
“You can’t buy the technology off the shelf,” Zenger points out. Furthermore, although the cost involved in sequencing is declining, making research applications more accessible, it currently remains too high for many aquaculturists – except for large commercial operations working with high-value species.
This, however, may be a temporary situation. Current research continues to form the basis for commercial applications, and costs continue to decline. Routine commercial use may not be that far beyond the horizon.
Source: The Fish Site






















