A Rapid, Sensitive RPA Assay for Detection of Vibrio Parahaemolyticus in Seafood
| Mon, 13 Apr 2020 - 11:14
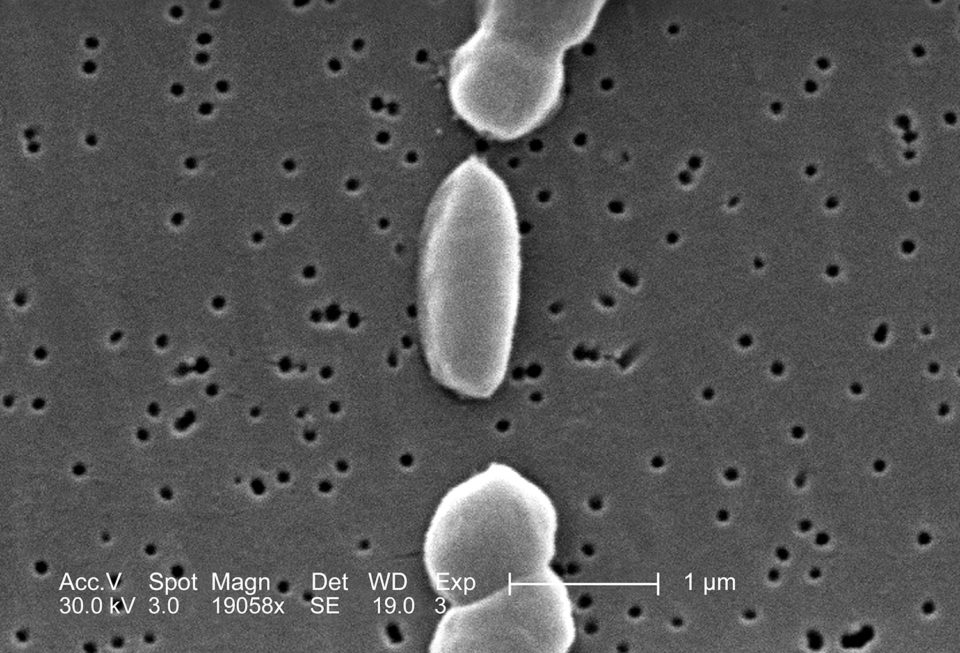
This scanning electron micrograph (SEM) depicts a number of Vibrio parahaemolyticusbacteria (Mag. 19058x), a serious pathogen in the seafood and other food industries. Photo by CDC/Janice Carr. Public domain.
The bacterium Vibrio parahaemolyticus is naturally present in brackish coastal environments and is frequently isolated from a variety of seafood. This bacterium is a major seafood-borne pathogen that causes gastrointestinal disorders due to ingestion of raw or undercooked seafood it contaminates. In recent years, outbreaks of V. parahaemolyticus infections have been a significant public health concern in many countries. Additionally, the number of V. parahaemolyticus infections has increased, and their reach has widened globally during recent years.
People infected with V. parahaemolyticus are characterized by an acute gastroenteritis disorder with clinical signs of diarrhea, headache and vomiting. Low immunity populations who become infected with V. parahaemolyticus may develop septicemia in severe cases. Controlling the amount of V. parahaemolyticus in seafood is an effective way to prevent infection by this pathogen; therefore, it is important to determine the levels of V. parahaemolyticus in seafood.
Early and rapid diagnosis is crucial for the management of V. parahaemolyticus infection. Different diagnostic methods for V. parahaemolyticus infection have been reported. Conventional culturing and immunological methods are common techniques used for V. parahaemolyticus detection. However, they are time-consuming and take a few days to provide a confirmed result after numerous analytical steps.
Advancements in biotechnology have led to the development of a number of gene amplification-based molecular detection technologies for V. parahaemolyticus, with varying degrees of sensitivity and specificity. These include polymerase chain reaction (PCR) and others. However, these tests can be impractical for on-site application due to the expensive instruments required and time-consuming operation. A simple, rapid, accurate and user-friendly platform is still needed for the early point-of-need (POD) detection of V. parahaemolyticus infection.
Recombinase polymerase amplification (RPA), a novel isothermal gene amplification technique, has been demonstrated to be a simple, rapid, specific, sensitive and cost-effective molecular assay to identify diverse pathogens. The RPA process for exponential amplification of the target sequence can be achieved in 20 minutes or less.
This article – adapted and summarized from the original publication [Geng, Y. et al. 2019. Development and evaluation of a rapid and sensitive RPA assay for specific detection of Vibrio parahaemolyticus in seafood. BMC Microbiol 19, 186 (2019) – describes a study to develop and evaluate a real-time RPA method for the simple and rapid detection of V. parahaemolyticus.
Study setup
A total of five V. parahaemolyticus strains, five other Vibrio species and 22 other bacterial strains were used to determine the specificity of the real-time RPA. For detailed information on the bacterial strains and their DNA extraction; RPA primers and probe; Real-time RPA reaction; real-time PCR for V. parahaemolyticus; specificity and analytical sensitivity analysis; evaluation with artificially contaminated samples; and statistical methods used, refer to the original publication.
This work was supported by the program of scientific research foundation in Universities of Hebei Province (QN2019025, Hebei, China); the General Administration of quality supervision, inspection and Quarantine of the People’s Republic of China (2016IK107, China); and the Biology postdoctoral Science of Hebei Normal University (183717, Hebei, China).
Results and discussion
The real-time RPA reaction was performed successfully at 38 degrees-C, and results were obtained within 20 minutes. The method only detected V. parahaemolyticus and did not show cross-reaction with other bacteria, exhibiting a high level of specificity. The study showed that the detection limit (LOD) of real-time RPA was 1.02 × 102 copies/reaction.
For artificially contaminated samples with different bacteria concentrations, V. parahaemolyticus could be detected within 5 to 12 min by real-time RPA in oyster sauce, codfish and sleeve-fish at concentrations as low as 4 Colony Forming Units (CFU)/25 g, 1 CFU/25 grams and 7 CFU/25 grams, respectively, after enrichment for 6 hours, but were detected in a minimum of 35 minutes by real-time PCR [with Ct (cycle threshold) values between 27 and 32; in a real time PCR assay, a positive reaction is detected by accumulation of a fluorescent signal. Ct the number of cycles required for the fluorescent signal to cross the threshold, to exceed the background level].
In recent years, diseases caused by food-borne pathogens have become a significant global public health issue. V. parahaemolyticus is one of the major pathogens causing food-borne illness, and contamination of food products with this pathogen has become a vital concern for food safety. Therefore, there was a need for rapid, specific and reliable diagnostic techniques that can be used effectively for better detection of V. parahaemolyticus in seafood, environmental and other various sample types.
In our study, we successfully established the real-time RPA assay targeting a specific gene of V. parahaemolyticus. Conventional RPA was successfully performed at 38 degrees-C and was completed within 20 minutes. The sensitivity of the real-time RPA assay was examined using serial dilutions of V. parahaemolyticus genomic DNA template. The limit of detection of the real-time RPA assay was 102 copies/reaction.
While the V. parahaemolyticus detection results by real-time RPA could be obtained in approximately 30 minutes, including the time for nucleic acid extraction, the reaction time for positive samples reached up to 1 hour when using real-time PCR. In addition, the real-time RPA assay was also successful in the detection of artificially contaminated seafood samples, and it performed better than real-time PCR with respect to detection speed.
For the real-time RPA assay described in this study, V. parahaemolyticus could be detected in artificially contaminated samples at concentrations as low as 1 CFU/25 grams within 12 minutes. Compared to other isothermal amplification techniques, RPA does not require initial heating for DNA denaturation and results can be obtained in less than 12 minutes which didn’t include the enrichment time. The real-time RPA assay has multiple advantages over other DNA amplification methods, including a quicker time-to-result for a single sample and others.
RPA has been widely explored for the molecular detection of diverse pathogens, and field testing has also been achieved for Dengue virus and avian influenza A virus infection. Moreover, the portable tube scanner used in the study, weighing only 1.75 kg with dimensions of 25 cm × 16.5 cm × 8.5 cm, is simpler than most real-time PCR machines and can be used in the field, running on battery power for an entire day.
Perspectives
In this study, we successfully developed a real-time RPA method based on an exo-probe for the detection of V. parahaemolyticus. With high sensitivity and specificity, the assay could be completed within 20 min and the approach is easy to perform in clinical settings without a requirement for sophisticated equipment, which renders it applicable at quarantine stations, ports of importation or sites of outbreaks.
The effective and rapid real-time RPA assay developed in this study would be highly useful in the monitoring of V. parahaemolyticus infection and has the potential to be a promising alternative to real-time PCR and other isothermal methods for rapidly testing V. parahaemolyticus infection, a significant risk in the seafood and other industries.
Source: Global Aquaculture Alliance